 under this model is fairly
small, and when set to zero in model II, found to be non-significant
(
under this model is fairly
small, and when set to zero in model II, found to be non-significant
(
In Table 9.2, we provide selected results from fitting the
following models: general sex-limitation (I); common effects
sex-limitation (II-IV); and scalar sex-limitation (V). We first note
that the general sex-limitation model provides a good fit to the data,
with ![]() . The estimate of
. The estimate of  under this model is fairly
small, and when set to zero in model II, found to be non-significant
(
under this model is fairly
small, and when set to zero in model II, found to be non-significant
(![]() = 2.54,
= 2.54, ![]() ). Thus, there is no evidence for
sex-specific additive genetic effects, and the common effects
sex-limitation model (model II) is favored over the general model. As
an exercise, the reader may wish to verify that the same conclusion is
reached if the general sex-limitation model with sex-specific dominant
genetic effects is compared to the common effects model with
). Thus, there is no evidence for
sex-specific additive genetic effects, and the common effects
sex-limitation model (model II) is favored over the general model. As
an exercise, the reader may wish to verify that the same conclusion is
reached if the general sex-limitation model with sex-specific dominant
genetic effects is compared to the common effects model with ![]() removed.
removed.
Note that under model II the dominant genetic parameter for females is
quite small; thus, when this parameter is fixed to zero in model III,
there is not a significant worsening of fit, and model III becomes the
most favored model. In model IV, we consider whether the dominant
genetic effect for males can also be fixed to zero. The
goodness-of-fit statistics indicate that this model fits the data
poorly (![]() ) and provides a significantly worse fit than model
III (
) and provides a significantly worse fit than model
III (![]() = 26.73,
= 26.73, ![]() ). Model IV is therefore
rejected and model III remains the favored one.
). Model IV is therefore
rejected and model III remains the favored one.
Finally, we consider the scalar sex-limitation model. Since there is
evidence for dominant genetic effects in males and not in females, it
seems unlikely that this model, which constrains the variance
components of females to be scalar multiples of the male variance
components, will provide a good fit to the data, unless the additive
genetic variance in females is also much smaller than the male
additive genetic variance. The model-fitting results support this
contention: the model provides a marginal fit to the data ( =
0.05), and is significantly worse than model II (![]() = 7.82,
= 7.82,
![]() ). We thus conclude from Table 9.2 that III is
the best fitting model. This conclusion would also be reached if AIC
was used to assess goodness-of-fit.
). We thus conclude from Table 9.2 that III is
the best fitting model. This conclusion would also be reached if AIC
was used to assess goodness-of-fit.
| MODEL | |||||
| Parameter | I | II | III | IV | V |
| 0.449 | 0.454 | 0.454 | 0.454 | 0.346 | |
| 0.172 | 0.000 | - | - | 0.288 | |
| 0.264 | 0.265 | 0.265 | 0.267 | 0.267 | |
| 0.210 | 0.240 | 0.240 | 0.342 | - | |
| 0.184 | 0.245 | 0.245 | - | - | |
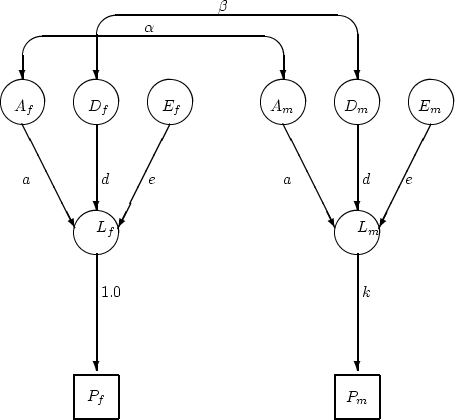 |
0.213 | 0.213 | 0.213 | 0.220 | - |
 |
0.198 | - | - | - | - |
| - | - | - | - | 0.778 | |
| 9.26 | 11.80 | 11.80 | 38.53 | 19.62 | |
| 8 | 9 | 10 | 11 | 11 | |
| 0.32 | 0.23 | 0.30 | 0.00 | 0.05 | |
| -6.74 | -6.20 | -8.20 | 16.53 | -2.38 | |
Using the parameter estimates under model III, the expected variance
of log BMI (residuals) in males and females can be calculated. A
little arithmetic reveals that the phenotypic variance of males is
markedly lower than that of females (0.17 vs. 0.28). Inspection of
the parameter estimates indicates that the sex difference in
phenotypic variance is due to increased genetic and environmental variance in females. However, the increase in
genetic variance in females is proportionately greater than the
increase in environmental variance, and this difference results in a
somewhat larger broad sense (i.e., ![]() ) heritability estimate
for females (75%) than for males (69%).
) heritability estimate
for females (75%) than for males (69%).