| First Twin |
Second Twin | |||||||||
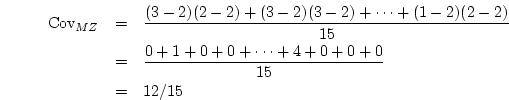 |
skew | kurt | skew | kurt | ||||||
| MZF | ||||||||||
| Young | 534 | .78 | 21.25 | 7.73 | 1.82 | 6.84 | 21.30 | 8.81 | 2.14 | 9.44 |
| Older | 637 | .69 | 23.11 | 11.87 | 1.22 | 2.53 | 22.97 | 11.25 | 1.08 | 2.11 |
| DZF | ||||||||||
| Young | 328 | .30 | 21.58 | 8.56 | 1.75 | 6.04 | 21.64 | 9.84 | 2.38 | 12.23 |
| Older | 380 | .32 | 22.77 | 10.93 | 1.40 | 4.03 | 22.95 | 12.63 | 1.26 | 2.43 |
| MZM | ||||||||||
| Young | 251 | .77 | 22.09 | 5.95 | 0.28 | 0.10 | 22.13 | 5.77 | 0.40 | 0.30 |
| Older | 281 | .70 | 24.22 | 6.42 | 0.11 | -0.05 | 24.30 | 7.85 | 0.43 | 0.63 |
| DZM | ||||||||||
| Young | 184 | .32 | 22.71 | 8.16 | 1.00 | 1.71 | 22.61 | 9.63 | 1.55 | 6.24 |
| Older | 137 | .37 | 24.18 | 8.28 | 0.41 | 0.70 | 24.08 | 7.42 | 0.72 | 0.43 |
| DZFM | ||||||||||
| Young | 464 | .23 | 21.33 | 6.89 | 1.06 | 1.84 | 22.47 | 6.81 | 0.76 | 1.72 |
| Older | 373 | .24 | 23.07 | 12.63 | 1.23 | 2.24 | 24.65 | 8.52 | 0.88 | 1.49 |
![[*]](crossref.png) ). Thus the fact that the like-sex DZ twin
correlations are less than one-half the size of the MZ correlations in
the young cohort suggests a contribution of genetic dominance, as well
as additive genetic variance, to individual differences in BMI.
Model-fitting analyses (e.g., []
are needed to determine whether the data:
). Thus the fact that the like-sex DZ twin
correlations are less than one-half the size of the MZ correlations in
the young cohort suggests a contribution of genetic dominance, as well
as additive genetic variance, to individual differences in BMI.
Model-fitting analyses (e.g., []
are needed to determine whether the data:
![[*]](footnote.png) separately for each like-sex twin group. These are summarized in
Table 6.2. If the joint distribution of twin pairs for
BMI is bivariate normal, these regressions should be non-significant.
Here, however, we observe a highly significant regression: on average,
pairs with high BMI values also exhibit larger intra-pair differences
in BMI. This is likely to be an artefact of scale, since using a
log-transformation substantially reduces the magnitude of the
polynomial regression (as well as reducing marginal measures of
skewness and kurtosis).
separately for each like-sex twin group. These are summarized in
Table 6.2. If the joint distribution of twin pairs for
BMI is bivariate normal, these regressions should be non-significant.
Here, however, we observe a highly significant regression: on average,
pairs with high BMI values also exhibit larger intra-pair differences
in BMI. This is likely to be an artefact of scale, since using a
log-transformation substantially reduces the magnitude of the
polynomial regression (as well as reducing marginal measures of
skewness and kurtosis).